A team of Australian and Canadian researchers has identified a major reason why some men are diagnosed with a very aggressive form of prostate cancer.
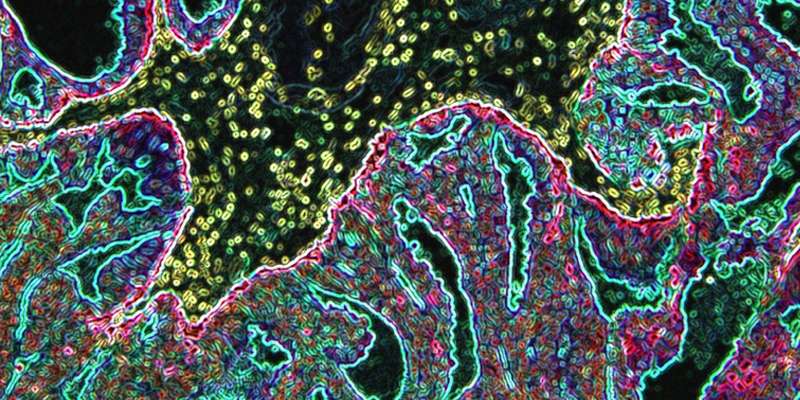
Severe prostate cancer is often also deviously difficult to predict. However, new findings reveal the genetic cause of why this is the case and might also make diagnosis easier in the future.
The researchers looked at changes in a known gene, the hereditary BRCA2, which leads to instability in the genome. This instability leads to faster development of prostate cancer and an increased risk of metastasis formation.
The study was done on a small group of men and the clearly shows that the genetic changes that occur as the result of mutations in the BRCA2 gene, results in the activation of previously known so-called oncogenes, which explains why certain men are developing rapid tumor progression.
Oncogenes are a type of gene that leads to improper protein production and this lack of function facilitates the creation of tumor cells. Most normal cells will undergo a programmed form of rapid cell death (apoptosis) when critical functions are altered. However, activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead.
The director of Monash University Partners Comprehensive Cancer Consortium and Monash BDI, lead author Professor Gail Risbridger said to Science Daily;
“This study shows how different these tumours are from ‘regular’ tumours and emphasises the importance of men knowing if they have a family history of prostate, breast or ovarian cancer in their family and may carry the BRCA2 gene fault,”
The study was led by Monash University’s Biomedicine Discovery Institute involving a consortium of Melbourne and Toronto researchers and clinician and has been published in Nature Communications and demonstrates the benefit of early diagnosis among those men who are at risk of developing aggressive prostate cancer.
Reference:
Renea A. Taylor et al “Germline BRCA2 mutations drive prostate cancer with distinct evolutionary trajectories”
Nature Communications January 9, 2017, DOI: 10.1038 / ncomms13671







![OpenAI. (2025). ChatGPT [Large language model]. https://chatgpt.com](https://www.illustratedcuriosity.com/files/media/55136/b1b0b614-5b72-486c-901d-ff244549d67a-350x260.webp)
![OpenAI. (2025). ChatGPT [Large language model]. https://chatgpt.com](https://www.illustratedcuriosity.com/files/media/55124/79bc18fa-f616-4951-856f-cc724ad5d497-350x260.webp)
![OpenAI. (2025). ChatGPT [Large language model]. https://chatgpt.com](https://www.illustratedcuriosity.com/files/media/55099/2638a982-b4de-4913-8a1c-1479df352bf3-350x260.webp)








